Structural Basis of Chemokine Sequestration by a Herpesvirus Decoy Receptor
Alexander, J.M., Nelson, C.A., van Berkel, V., Lau, E.K., Studts, J.M., Brett, T.J., Speck, S.H., Handel, T.M., Virgin, H.W., Fremont, D.H.(2002) Cell 111: 343-356
- PubMed: 12419245
- DOI: https://doi.org/10.1016/s0092-8674(02)01007-3
- Primary Citation of Related Structures:
1MKF, 1ML0 - PubMed Abstract:
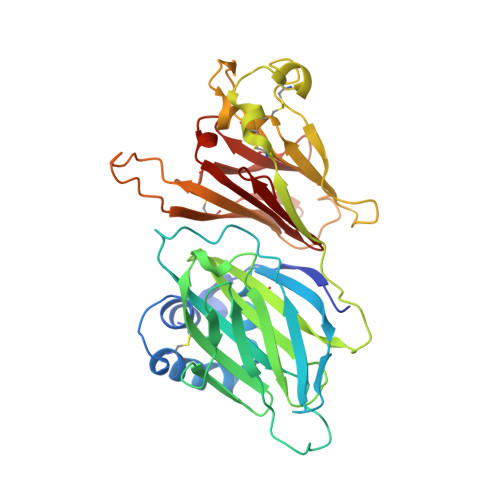
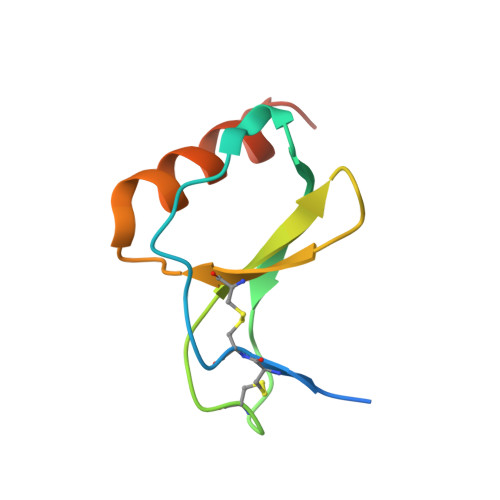
The M3 protein encoded by murine gamma herpesvirus68 (gamma HV68) functions as an immune system saboteur by the engagement of chemoattractant cytokines, thereby altering host antiviral inflammatory responses. Here we report the crystal structures of M3 both alone and in complex with the CC chemokine MCP-1. M3 is a two-domain beta sandwich protein with a unique sequence and topology, forming a tightly packed anti-parallel dimer. The stoichiometry of the MCP-1:M3 complex is 2:2, with two monomeric chemokines embedded at distal ends of the preassociated M3 dimer. Conformational flexibility and electrostatic complementation are both used by M3 to achieve high-affinity and broad-spectrum chemokine engagement. M3 also employs structural mimicry to promiscuously sequester chemokines, engaging conservative structural elements associated with both chemokine homodimerization and binding to G protein-coupled receptors.
Organizational Affiliation:
Department of Pathology and Immunology, Washington University School of Medicine, 660 South Euclid Avenue, St. Louis, MO 63110, USA.