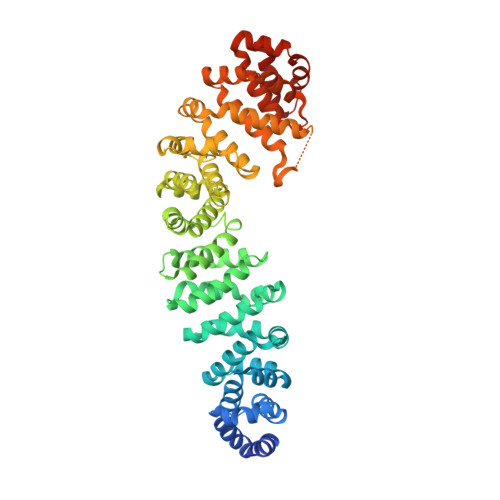
Structure of a human Tcf4-beta-catenin complex.
Poy, F., Lepourcelet, M., Shivdasani, R.A., Eck, M.J.(2001) Nat Struct Biol 8: 1053-1057
- PubMed: 11713476
- DOI: https://doi.org/10.1038/nsb720
- Primary Citation of Related Structures:
1JPW - PubMed Abstract:
The multifunctional protein beta-catenin is important for cell adhesion, because it binds cadherins, and the Wnt signal transduction pathway, where it interacts with the Adenomatous polyposis coli (APC) protein and TCF/Lef family transcription factors. Mutations in APC or in beta-catenin are estimated to trigger formation of over 90% of all colon cancers. In colonic epithelia, these mutations produce elevated levels of Tcf4-beta-catenin, which stimulates a transcriptional response that initiates polyp formation and eventually malignant growth. Thus, disruption of the Tcf4-beta-catenin interaction may be an attractive goal for therapeutic intervention. Here we describe the crystal structure of a human Tcf4-beta-catenin complex and compare it with recent structures of beta-catenin in complex with Xenopus Tcf3 (XTcf3) and mammalian E-cadherin. The structure reveals anticipated similarities with the closely related XTcf3 complex but unexpectedly lacks one component observed in the XTcf3 structure.
Organizational Affiliation:
Department of Cancer Biology, Dana-Farber Cancer Institute, 44 Binney Street, Boston, Massachusetts 02115, USA.