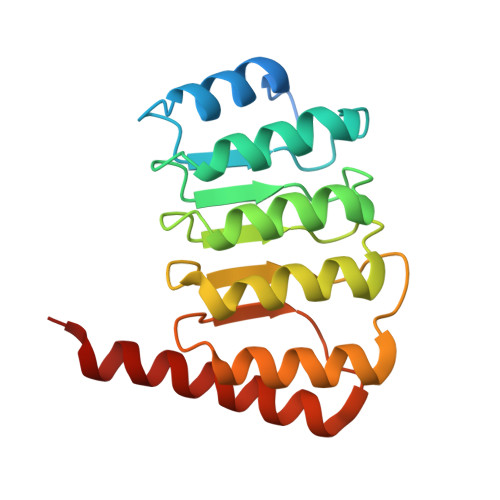
Crystal structure of the C-terminal half of tropomodulin and structural basis of actin filament pointed-end capping.
Krieger, I., Kostyukova, A., Yamashita, A., Nitanai, Y., Maeda, Y.(2002) Biophys J 83: 2716-2725
- PubMed: 12414704
- DOI: https://doi.org/10.1016/S0006-3495(02)75281-8
- Primary Citation of Related Structures:
1IO0 - PubMed Abstract:
Tropomodulin is the unique pointed-end capping protein of the actin-tropomyosin filament. By blocking elongation and depolymerization, tropomodulin regulates the architecture and the dynamics of the filament. Here we report the crystal structure at 1.45-A resolution of the C-terminal half of tropomodulin (C20), the actin-binding moiety of tropomodulin. C20 is a leucine-rich repeat domain, and this is the first actin-associated protein with a leucine-rich repeat. Binding assays suggested that C20 also interacts with the N-terminal fragment, M1-M2-M3, of nebulin. Based on the crystal structure, we propose a model for C20 docking to the actin subunit at the pointed end. Although speculative, the model is consistent with the idea that a tropomodulin molecule competes with an actin subunit for a pointed end. The model also suggests that interactions with tropomyosin, actin, and nebulin are all possible sources of influences on the dynamic properties of pointed-end capping by tropomodulin.
Organizational Affiliation:
Laboratory for Structural Biochemistry, RIKEN Harima Institute at SPring-8, Mikazuki, Sayo, Hyogo, Japan 679-5148.