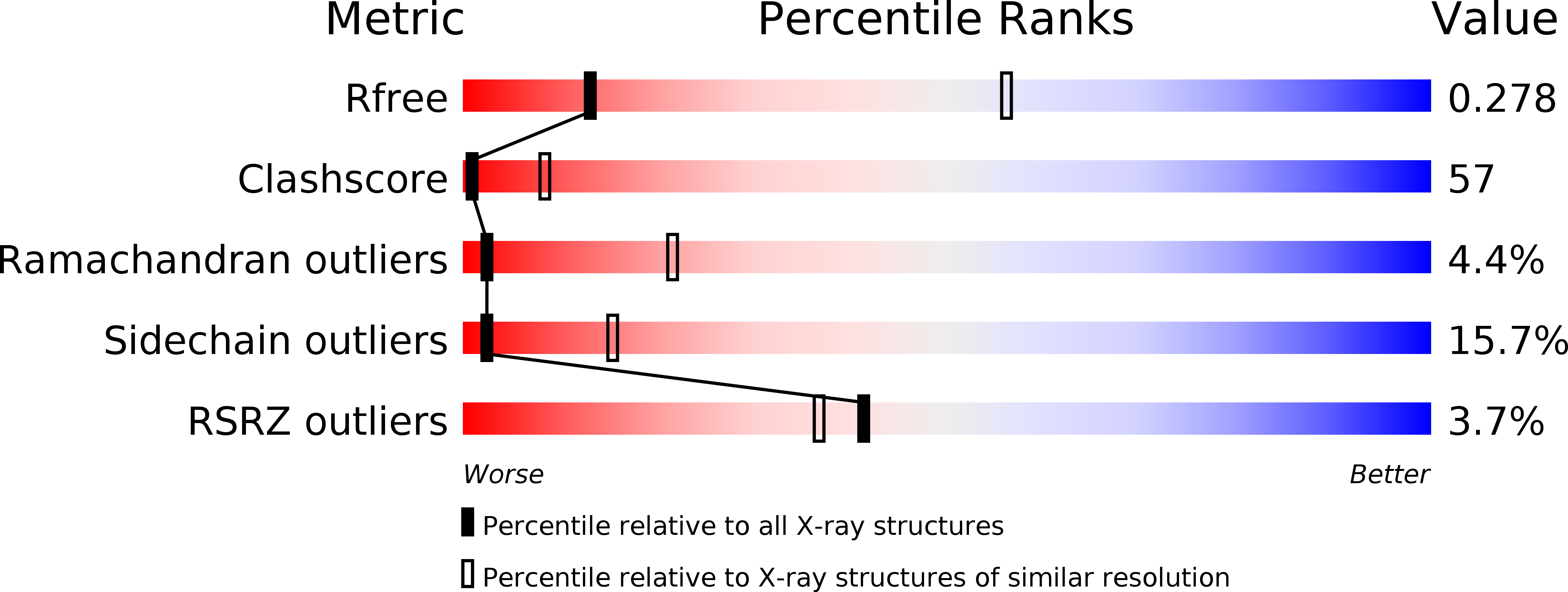
Structure of human muscle creatine kinase.
Shen, Y.Q., Tang, L., Zhou, H.M., Lin, Z.J.(2001) Acta Crystallogr D Biol Crystallogr 57: 1196-1200
- PubMed: 11517911
- DOI: https://doi.org/10.1107/s0907444901007703
- Primary Citation of Related Structures:
1I0E - PubMed Abstract:
The crystal structure of human muscle creatine kinase has been determined by the molecular-replacement method and refined at 3.5A resolution. The structures of both the monomer and the dimer closely resemble those of the other known structures in the creatine kinase family. Two types of dimers, one with a non-crystallographic twofold symmetry axis and the other with a crystallographic twofold symmetry axis, were found to exist simultaneously in the crystal. These dimers form an infinite "double-helix"-like structure along an unusual long crystallographic 3(1) axis.
Organizational Affiliation:
National Laboratory of Biological Macromolecules, Institute of Biophysics, Academica Sinica, Beijing 100101, People's Republic of China.