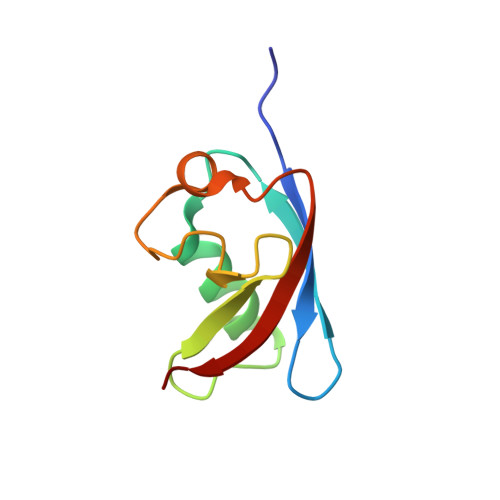
The Ubx Domain: A Widespread Ubiquitin-Like Module
Buchberger, A., Howard, M.J., Proctor, M., Bycroft, M.M.(2001) J Mol Biol 307: 17
- PubMed: 11243799
- DOI: https://doi.org/10.1006/jmbi.2000.4462
- Primary Citation of Related Structures:
1H8C - PubMed Abstract:
The UBX domain is an 80 amino acid residue module that is present typically at the carboxyl terminus of a variety of eukaryotic proteins. In an effort to elucidate the function of UBX domains, we solved the three-dimensional structure of the UBX domain of human Fas-associated factor-1 (FAF1) by NMR spectroscopy. The structure has a beta-Grasp fold characterised by a beta-beta-alpha-beta-beta-alpha-beta secondary-structure organisation. The five beta strands are arranged into a mixed sheet in the order 21534. The longer first helix packs across the first three strands of the sheet, and a second shorter 3(10) helix is located in an extended loop connecting strands 4 and 5. In the absence of significant sequence similarity, the UBX domain can be superimposed with ubiquitin with an r.m.s.d. of 1.9 A, suggesting that the two structures share the same superfold, and an evolutionary relationship. However, the absence of a carboxyl-terminal extension containing a double glycine motif and of suitably positioned lysine side-chains makes it highly unlikely that UBX domains are either conjugated to other proteins or part of mixed UBX-ubiquitin chains. Database searches revealed that most UBX domain-containing proteins belong to one of four evolutionarily conserved families represented by the human FAF1, p47, Y33K, and Rep8 proteins. A role of the UBX domain in ubiquitin-related processes is suggested.
Organizational Affiliation:
MRC Centre for Protein Engineering, Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge CB2 1EW, UK.