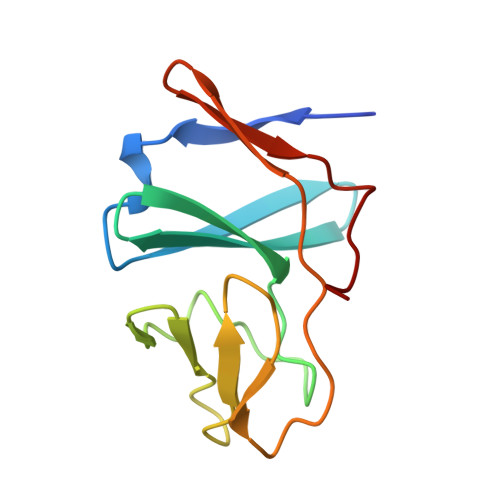
A cluster exposed: structure of the Rieske ferredoxin from biphenyl dioxygenase and the redox properties of Rieske Fe-S proteins.
Colbert, C.L., Couture, M.M., Eltis, L.D., Bolin, J.T.(2000) Structure 8: 1267-1278
- PubMed: 11188691
- DOI: https://doi.org/10.1016/s0969-2126(00)00536-0
- Primary Citation of Related Structures:
1FQT - PubMed Abstract:
Ring-hydroxylating dioxygenases are multicomponent systems that initiate biodegradation of aromatic compounds. Many dioxygenase systems include Rieske-type ferredoxins with amino acid sequences and redox properties remarkably different from the Rieske proteins of proton-translocating respiratory and photosynthetic complexes. In the latter, the [Fe2S2] clusters lie near the protein surface, operate at potentials above +300 mV at pH 7, and express pH- and ionic strength-dependent redox behavior. The reduction potentials of the dioxygenase ferredoxins are approximately 150 mV and are pH-independent. These distinctions were predicted to arise from differences in the exposure of the cluster and/or interactions of the histidine ligands.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907, USA.