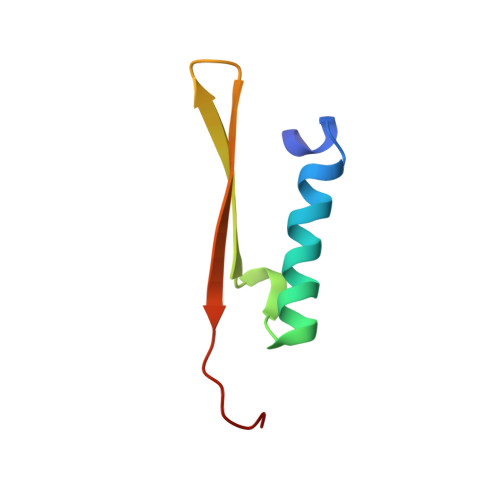
Structural basis for the topological specificity function of MinE.
King, G.F., Shih, Y.L., Maciejewski, M.W., Bains, N.P., Pan, B., Rowland, S.L., Mullen, G.P., Rothfield, L.I.(2000) Nat Struct Biol 7: 1013-1017
- PubMed: 11062554
- DOI: https://doi.org/10.1038/80917
- Primary Citation of Related Structures:
1EV0 - PubMed Abstract:
Correct positioning of the division septum in Escherichia coli depends on the coordinated action of the MinC, MinD and MinE proteins. Topological specificity is conferred on the MinCD division inhibitor by MinE, which counters MinCD activity only in the vicinity of the preferred midcell division site. Here we report the structure of the homodimeric topological specificity domain of Escherichia coli MinE and show that it forms a novel alphabeta sandwich. Structure-directed mutagenesis of conserved surface residues has enabled us to identify a spatially restricted site on the surface of the protein that is critical for the topological specificity function of MinE.
Organizational Affiliation:
Department of Biochemistry, University of Connecticut Health Center, 263 Farmington Avenue, Farmington, Connecticut 06032, USA. glenn@psel.uchc.edu