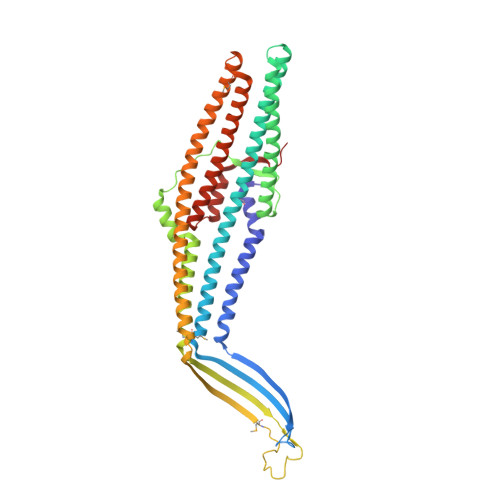
Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export.
Koronakis, V., Sharff, A., Koronakis, E., Luisi, B., Hughes, C.(2000) Nature 405: 914-919
- PubMed: 10879525
- DOI: https://doi.org/10.1038/35016007
- Primary Citation of Related Structures:
1EK9 - PubMed Abstract:
Diverse molecules, from small antibacterial drugs to large protein toxins, are exported directly across both cell membranes of gram-negative bacteria. This export is brought about by the reversible interaction of substrate-specific inner-membrane proteins with an outer-membrane protein of the TolC family, thus bypassing the intervening periplasm. Here we report the 2.1-A crystal structure of TolC from Escherichia coli, revealing a distinctive and previously unknown fold. Three TolC protomers assemble to form a continuous, solvent-accessible conduit--a 'channel-tunnel' over 140 A long that spans both the outer membrane and periplasmic space. The periplasmic or proximal end of the tunnel is sealed by sets of coiled helices. We suggest these could be untwisted by an allosteric mechanism, mediated by protein-protein interactions, to open the tunnel. The structure provides an explanation of how the cell cytosol is connected to the external environment during export, and suggests a general mechanism for the action of bacterial efflux pumps.
Organizational Affiliation:
Department of Pathology, University of Cambridge, UK.