Three-dimensional structure analysis of mu-agatoxins: further evidence for common motifs among neurotoxins with diverse ion channel specificities.
Omecinsky, D.O., Holub, K.E., Adams, M.E., Reily, M.D.(1996) Biochemistry 35: 2836-2844
- PubMed: 8608119
- DOI: https://doi.org/10.1021/bi952605r
- Primary Citation of Related Structures:
1EIT - PubMed Abstract:
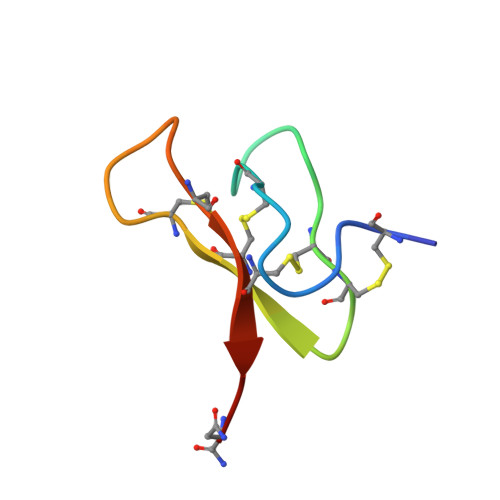
We report the solution structure of mu-agatoxin-I (mu-Aga-I) and model structures of the closely related mu-agatoxin-IV (mu-Aga-IV) which were isolated from venom of the American funnel web spider, Agelenopsis aperta. These toxins, which modify the kinetics of neuronal voltage-activated sodium channels in insects, are C-terminally amidated peptides composed to 36 amino acids, including four internal disulfide bonds. The structure of mu-Aga-I was determined by NMR and distance geometry/molecular dynamics calculations. Structural calculations were carried out using 256 interresidue NOE-derived distance restraints and 25 angle restraints obtained from vicinal coupling constants. The peptide contains eight cysteines involved in disulfide bonds, the pairings of which were uncertain and had to be determined from preliminary structure calculations. The toxin has an average rmsd of 0.89 A for the backbone atoms among 38 converged conformers. The structure consists of a well-defined triple-stranded beta-sheet involving residues 7-9, 20-24, and 30-34 and four tight turns. A homologous peptide, mu-Aga-IV, exhibited two distinct and equally populated conformations in solution, which complicated spectral analysis. Analysis of sequential NOE's confirmed that the conformers arose from cis and trans peptide bonds involving a proline at position 15. Models were developed for both conformers based on the mu-Aga-I structure. Our structural data show that the mu-agatoxins, although specific modifiers of sodium channels, share common secondary and tertiary structural motifs with phylogenetically diverse peptide toxins targeting a variety of channel types. The mu-agatoxins add voltage-sensitive sodium channel activity to a growing list of neurotoxic effects elicited by peptide toxins which share the same global fold yet differ in their animal origin and ion channel selectivity.
Organizational Affiliation:
Department of Chemistry, Parke-Davis Pharmaceutical Research Division of Warner Lambert Company, Ann Arbor, Michigan 48105, USA.