Solution Structure, Hydrodynamics and Thermodynamics of the Uvrb C-Terminal Domain.
Alexandrovich, A.A., Czisch, M.M., Frenkiel, T.T., Kelly, G.G., Goosen, N.N., Moolenaar, G.G., Chowdhry, B.B., Sanderson, M.M., Lane, A.A.(2001) J Biomol Struct Dyn 19: 219-236
- PubMed: 11697728
- DOI: https://doi.org/10.1080/07391102.2001.10506734
- Primary Citation of Related Structures:
1E52 - PubMed Abstract:
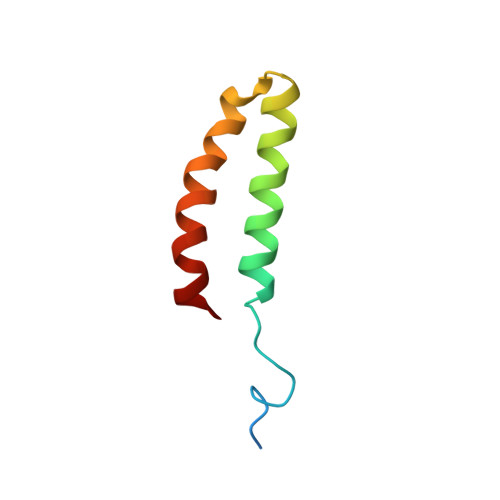
The solution structure, thermodynamic stability and hydrodynamic properties of the 55-residue C-terminal domain of UvrB that interacts with UvrC during excision repair in E. coli have been determined using a combination of high resolution NMR, ultracentrifugation, 15N NMR relaxation, gel permeation, NMR diffusion, circular dichroism and differential scanning calorimetry. The subunit molecular weight is 7,438 kDa., compared with 14.5+/-1.0 kDa. determined by equilibrium sedimentation, indicating a dimeric structure. The structure determined from NMR showed a stable dimer of anti-parallel helical hairpins that associate in an unusual manner, with a small and hydrophobic interface. The Stokes radius of the protein decreases from a high plateau value (ca. 22 A) at protein concentrations greater than 4 microM to about 18 A at concentrations less than 0.1 microM. The concentration and temperature-dependence of the far UV circular dichroism show that the protein is thermally stable (Tm ca. 71.5 degrees C at 36 microM). The simplest model consistent with these data was a dimer dissociating into folded monomers that then unfolds co-operatively. The van't Hoff enthalpy and dissociation constant for both transition was derived by fitting, with deltaH1=23 kJ mol(-1). K1(298)=0.4 microM and deltaH2= 184 kJ mol(-1). This is in good agreement with direct calorimetric analysis of the thermal unfolding of the protein, which gave a calorimetric enthalpy change of 181 kJ mol(-1) and a van't Hoff enthalpy change of 354 kJ mol(-1), confirming the dimer to monomer unfolding. The thermodynamic data can be reconciled with the observed mode of dimerisation. 15N NMR relaxation measurements at 14.1 T and 11.75 T confirmed that the protein behaves as an asymmetric dimer at mM concentrations, with a flexible N-terminal linker for attachment to the remainder of the UvrB protein. The role of dimerisation of this domain in the excision repair mechanism is discussed.
Organizational Affiliation:
Randall Centre for Molecular Mechanisms of Cell Function, New Hunt's House, King's College, London, UK.