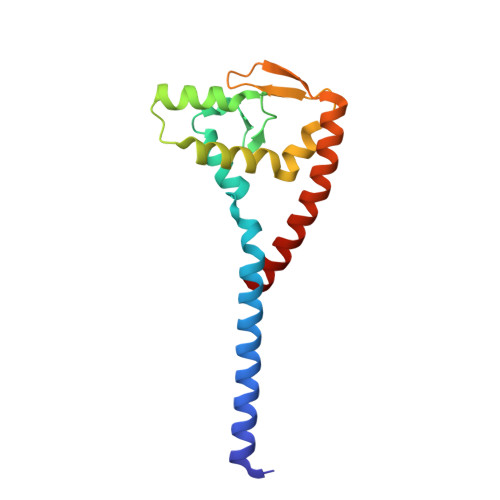
Crystal structure of the bacterial conjugation repressor finO.
Ghetu, A.F., Gubbins, M.J., Frost, L.S., Glover, J.N.(2000) Nat Struct Biol 7: 565-569
- PubMed: 10876242
- DOI: https://doi.org/10.1038/76790
- Primary Citation of Related Structures:
1DVO - PubMed Abstract:
The conjugative transfer of F-like plasmids is repressed by FinO, an RNA binding protein. FinO interacts with the F-plasmid encoded traJ mRNA and its antisense RNA, FinP, stabilizing FinP against endonucleolytic degradation and facilitating sense-antisense RNA recognition. Here we present the 2.0 A resolution X-ray crystal structure of FinO, lacking its flexible N-terminal extension. FinO adopts a novel, elongated, largely helical conformation. An N-terminal region, previously shown to contact RNA, forms a positively charged alpha-helix (helix 1) that protrudes 45 A from the central core of FinO. A C-terminal region of FinO that is implicated in RNA interactions also extends out from the central body of the protein, adopting a helical conformation and packing against the base of the N-terminal helix. A highly positively charged patch on the surface of the FinO core may present another RNA binding surface. The results of an in vitro RNA duplexing assay demonstrate that the flexible N-terminal region of FinO plays a key role in FinP-traJ RNA recognition, and supports our proposal that this region and the N-terminus of helix 1 interact with and stabilize paired, complementary RNA loops in a kissing complex.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, Edmonton, Alberta, T6G 2H7, Canada.