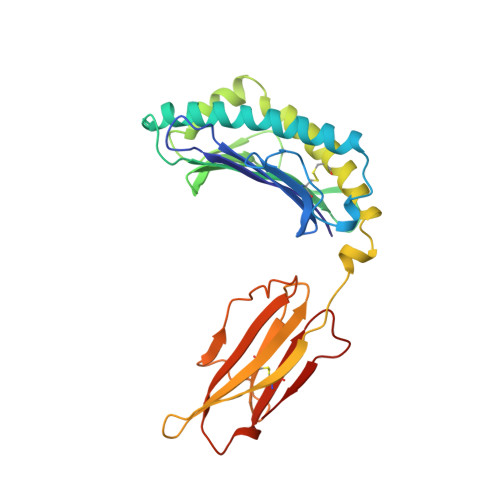
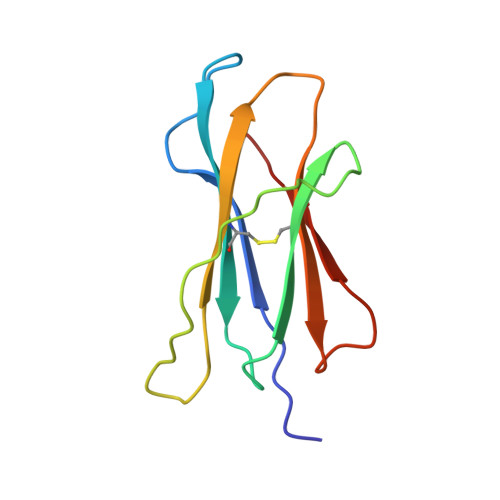
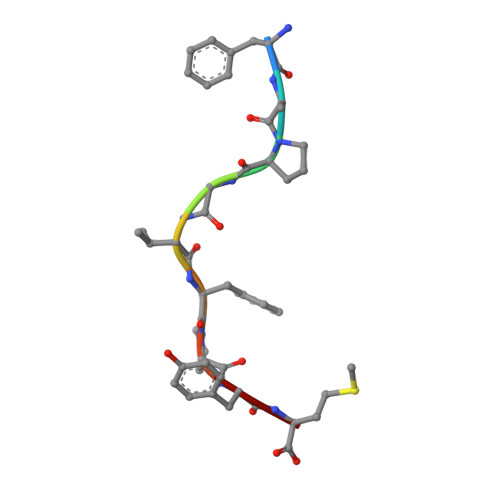
Structural evidence of T cell xeno-reactivity in the absence of molecular mimicry.
Zhao, R., Loftus, D.J., Appella, E., Collins, E.J.(1999) J Exp Med 189: 359-370
- PubMed: 9892618
- DOI: https://doi.org/10.1084/jem.189.2.359
- Primary Citation of Related Structures:
1B0G, 1BZ9 - PubMed Abstract:
The T cell receptor (TCR), from a xeno-reactive murine cytotoxic T lymphocyte clone AHIII12.2, recognizes murine H-2Db complexed with peptide p1027 (FAPGVFPYM), as well as human HLA-A2.1 complexed with peptide p1049 (ALWGFFPVL). A commonly proposed model (the molecular mimicry model) used to explain TCR cross-reactivity suggests that the molecular surfaces of the recognized complexes are similar in shape, charge, or both, in spite of the primary sequence differences. To examine the mechanism of xeno-reactivity of AHIII12.2, we have determined the crystal structures of A2/p1049 and Db/p1027 to 2.5 A and 2.8 A resolution, respectively. The crystal structures show that the TCR footprint regions of the two class I complexes are significantly different in shape and charge. We propose that rather than simple molecular mimicry, unpredictable arrays of common and differential contacts on the two class I complexes are used for their recognition by the same TCR.
Organizational Affiliation:
Department of Microbiology and Immunology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.