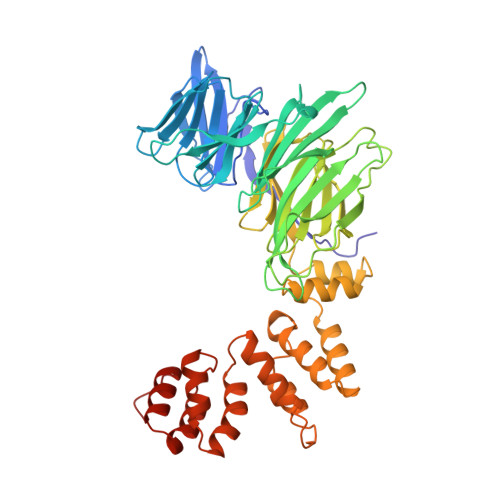
Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker.
Haar, E.T., Musacchio, A., Harrison, S.C., Kirchhausen, T.(1998) Cell 95: 563-573
- PubMed: 9827808
- DOI: https://doi.org/10.1016/s0092-8674(00)81623-2
- Primary Citation of Related Structures:
1BPO - PubMed Abstract:
Clathrin triskelions form the lattice that organizes recruitment of proteins to coated pits and helps drive vesiculation of the lipid bilayer. We report the crystal structure at 2.6 A resolution of a 55 kDa N-terminal fragment from the 190 kDa clathrin heavy chain. The structure comprises the globular "terminal domain" and the linker that joins it to the end of a triskelion leg. The terminal domain is a seven-blade beta propeller, a structure well adapted to interaction with multiple partners, such as the AP-1 and AP-2 sorting adaptor complexes and the nonvisual arrestins. The linker is an alpha-helical zigzag emanating from the propeller domain. We propose that this simple motif may extend into the rest of the clathrin leg.
Organizational Affiliation:
Department of Cell Biology and Center for Blood Research, Harvard Medical School, Boston, Massachusetts 02115-5701, USA.