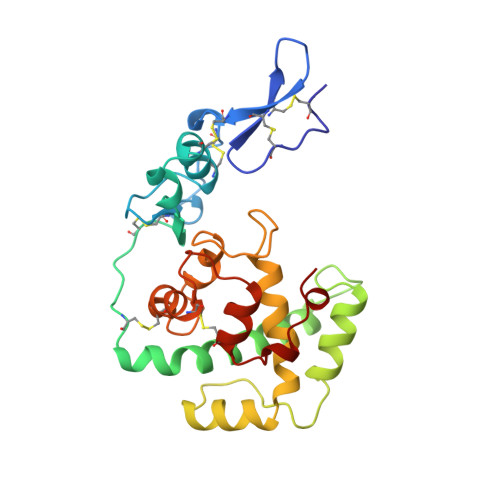
Crystal structure of a pair of follistatin-like and EF-hand calcium-binding domains in BM-40.
Hohenester, E., Maurer, P., Timpl, R.(1997) EMBO J 16: 3778-3786
- PubMed: 9233787
- DOI: https://doi.org/10.1093/emboj/16.13.3778
- Primary Citation of Related Structures:
1BMO - PubMed Abstract:
BM-40 (also known as SPARC or osteonectin) is an anti-adhesive secreted glycoprotein involved in tissue remodelling. Apart from an acidic N-terminal segment, BM-40 consists of a follistatin-like (FS) domain and an EF-hand calcium-binding (EC) domain. Here we report the crystal structure at 3.1 A resolution of the FS-EC domain pair of human BM-40. The two distinct domains interact through a small interface that involves the EF-hand pair of the EC domain. Residues implicated in cell binding, inhibition of cell spreading and disassembly of focal adhesions cluster on one face of BM-40, opposite the binding epitope for collagens and the N-linked carbohydrate. The elongated FS domain is structurally related to serine protease inhibitors of the Kazal family. Notable differences are an insertion into the inhibitory loop in BM-40 and a protruding N-terminal beta-hairpin with striking similarities to epidermal growth factor. This hairpin is likely to act as a rigid spacer in proteins containing tandemly repeated FS domains, such as follistatin and agrin, and forms the heparin-binding site in follistatin.
Organizational Affiliation:
Department of Crystallography, Birkbeck College, London, UK.