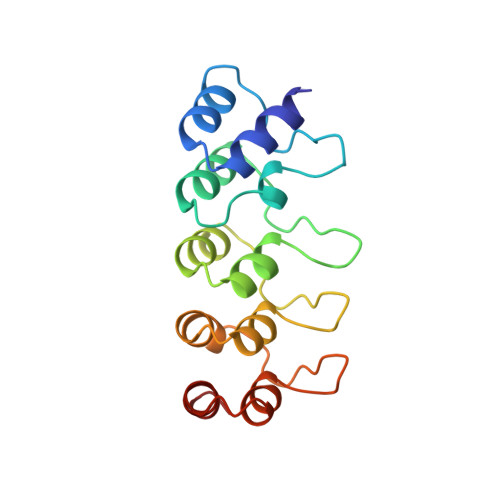
Structure of human cyclin-dependent kinase inhibitor p19INK4d: comparison to known ankyrin-repeat-containing structures and implications for the dysfunction of tumor suppressor p16INK4a.
Baumgartner, R., Fernandez-Catalan, C., Winoto, A., Huber, R., Engh, R.A., Holak, T.A.(1998) Structure 6: 1279-1290
- PubMed: 9782052
- DOI: https://doi.org/10.1016/s0969-2126(98)00128-2
- Primary Citation of Related Structures:
1BD8 - PubMed Abstract:
The four members of the INK4 gene family (p16(INK4a), p15(INK4b), p18(INK4c) and p19(INK4d)) inhibit the closely related cyclin-dependent kinases CDK4 and CDK6 as part of the regulation of the G1-->S transition in the cell-division cycle. Loss of INK4 gene product function, particularly that of p16(INK4a), is found in 10-60% of human tumors, suggesting that broadly applicable anticancer therapies might be based on restoration of p16(INK4a) CDK inhibitory function. Although much less frequent, defects of p19(INK4d) have also been associated with human cancer (osteosarcomas). The protein structures of some INK4 family members, determined by nuclear magnetic resonance (NMR) spectroscopy and X-ray techniques, have begun to clarify the functional role of p16(INK4a) and the dysfunction introduced by the mutations associated with human tumors.
Organizational Affiliation:
Max Planck Institute for Biochemistry D-82152, Martinsried, Federal Republic of Germany.