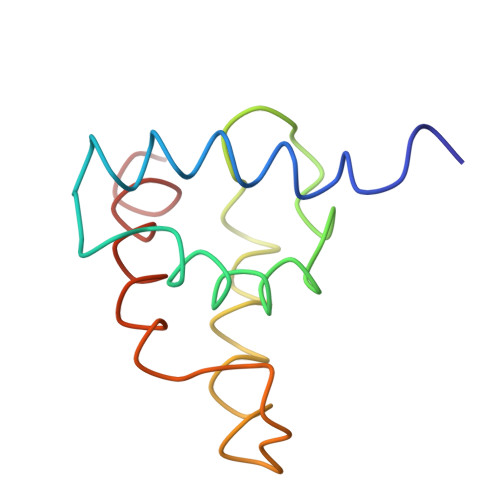
The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus, and implications for the morphology of retroviral assembly.
Conte, M.R., Klikova, M., Hunter, E., Ruml, T., Matthews, S.(1997) EMBO J 16: 5819-5826
- PubMed: 9312040
- DOI: https://doi.org/10.1093/emboj/16.19.5819
- Primary Citation of Related Structures:
1BAX - PubMed Abstract:
The Mason-Pfizer monkey virus (M-PMV) is the prototype of the type D retroviruses. In type B and D retroviruses, the Gag protein pre-assembles before association with the membrane, whereas in type C retroviruses (lentiviruses, BLV/HTLV group) Gag is targeted efficiently to the plasma membrane, where the particle formation occurs. The N-terminal domain of Gag, the matrix protein (MA), plays a critical role in determining this morphogenic difference. We have determined the three-dimensional solution structure of the M-PMV MA by heteronuclear nuclear magnetic resonance. The protein contains four alpha-helices that are structurally similar to the known type C MA structures. This similarity implies possible common assembly units and membrane-binding mechanisms for type C and B/D retroviruses. In addition to this, the interpretation of mutagenesis data has enabled us to identify, for the first time, the structural basis of a putative intracellular targeting motif.
Organizational Affiliation:
Department of Biochemistry, Imperial College of Science, Technology and Medicine, University of London, London SW7 2AY, UK.