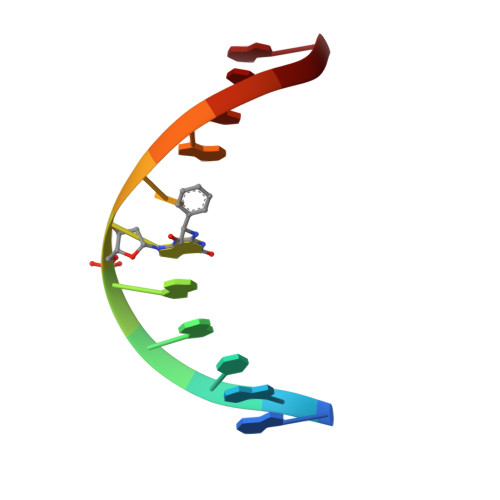
Styrene oxide adducts in an oligodeoxynucleotide containing the human N-ras codon 12 sequence: structural refinement of the minor groove R(12,2)- and S(12,2)-alpha-(N2-guanyl) stereoisomers from 1H NMR.
Zegar, I.S., Setayesh, F.R., DeCorte, B.L., Harris, C.M., Harris, T.M., Stone, M.P.(1996) Biochemistry 35: 4334-4348
- PubMed: 8605182
- DOI: https://doi.org/10.1021/bi952086s
- Primary Citation of Related Structures:
1AF1 - PubMed Abstract:
The structures of the (R)- and (S)-alpha-(N2-guanyl)styrene oxide adducts at X6 in d(GGCAGXTGGTG).d(CACCACCTGCC), encompassing codon 12 of the human n-ras protooncogene (underlined), were refined from 1H NMR data. These were the R(12,2) and S(12,2) adducts. For the R(12,2) adduct, upfield chemical shifts were observed for the T7 H6, H1', and N3H resonances. At 30 degrees C, R-SOG 6 N1H, T7 N3H, and T10 N3H disappeared due to exchange with solvent. For the S(12,2) adduct, S-SOG6 H1' shifted upfield 0.33 ppm, but all imino resonances were observed. The styrene methylene protons were nonequivalent for both adducts, suggesting hydrogen bonding between the hydroxyl and C18 O2 or O4' in the R(12,2) adduct and C17 O2 in the S(12,2) adduct. The styrene aromatic protons appeared as three signals in the R(12,2) adduct and as two signals in the S(12,2) adduct, suggesting rapid rotation of the styrene ring on the NMR time scale. NOE data revealed that the phenyl ring was oriented in the 3'-direction relative to R-SOG6 for the R(12,2) adduct and in the 5'-direction relative to S-SOG6 for the S(12,2) adduct. A total of 253 and 221 interproton distances were obtained from relaxation matrix analyses of the R(12,2) and S(12,2) adducts, respectively. NOE-restrained molecular dynamics calculations converged with root mean square deviations of 0.8-1.2 A for the R(12,2) adduct and 0.82-1.4 A for the S(12,2) adduct. Complete relaxation matrix analyses of the nine inner base pairs yielded sixth root residual indices between calculated and experimental NOE intensities of 8.8 x 10(-2) for the R(12,2) adduct and 7.9 x 10(-2) for the S(12,2) adduct. The refined structure for the R(12,2) adduct showed a 0.4 A increase in the stretch of R-SOG6.C17 and T7.A16, and a 1-2 A widening of the minor groove at and adjacent to the SO lesion, with the styrene ring oriented edgewise in the minor groove. Smaller minor groove disturbances were observed for the S(12,2) adduct, which had the styrene ring oriented flat in the minor groove. No DNA bending was predicted by the calculated structures.
Organizational Affiliation:
Center for Molecular Toxicology, Vanderbilt University, Nashville, Tennessee 37235, USA.