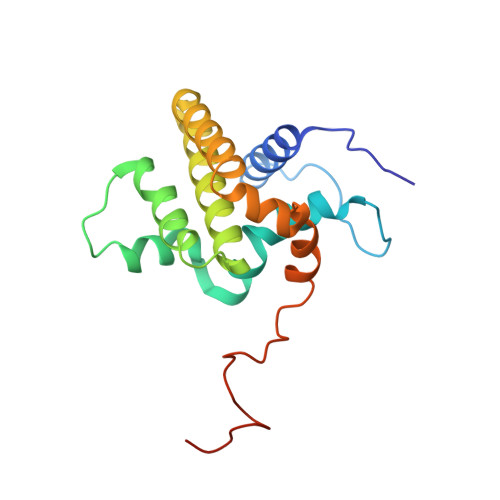
Structural Model of the BCL-w-BID Peptide Complex and Its Interactions with Phospholipid Micelles.
Denisov, A.Y., Chen, G., Sprules, T., Moldoveanu, T., Beauparlant, P., Gehring, K.(2006) Biochemistry 45: 2250-2256
- PubMed: 16475813
- DOI: https://doi.org/10.1021/bi052332s
- Primary Citation of Related Structures:
1ZY3 - PubMed Abstract:
A peptide corresponding to the BH3 region of the proapoptotic protein, BID, could be bound in the cleft of the antiapoptotic protein, BCL-w. This binding induced major conformational rearrangements in both the peptide and protein components of the complex and led to the displacement and unfolding of the BCL-w C-terminal alpha-helix. The structure of BCL-w with a bound BID-BH3 peptide was determined using NMR spectroscopy and molecular docking. These studies confirmed that a region of 16 residues of the BID-BH3 peptide is responsible for its strong binding to BCL-w and BCL-x(L). The interactions of BCL-w and the BID-BH3 peptide complex with dodecylphosphocholine micelles were characterized and showed that the conformational change of BCL-w upon lipid binding occurred at the same time as the release and unfolding of the BH3 peptide.
Organizational Affiliation:
Department of Biochemistry, McGill University, Montreal, Quebec H3G 1Y6, Canada.