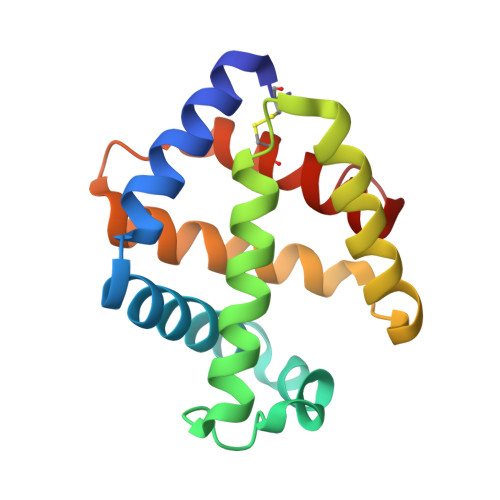
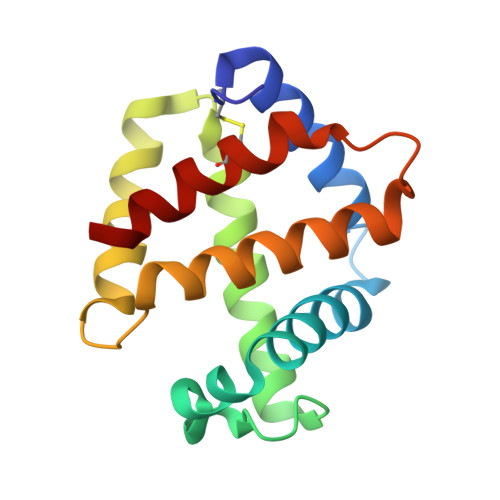
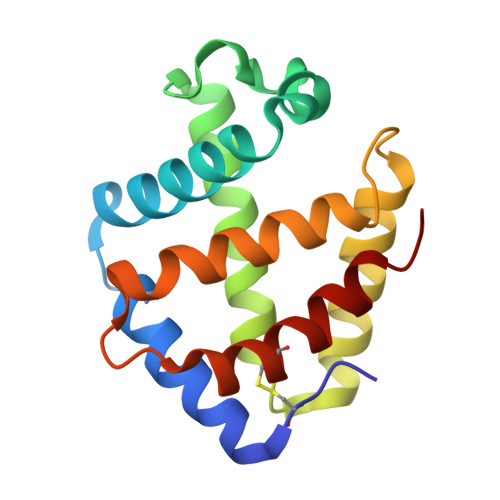
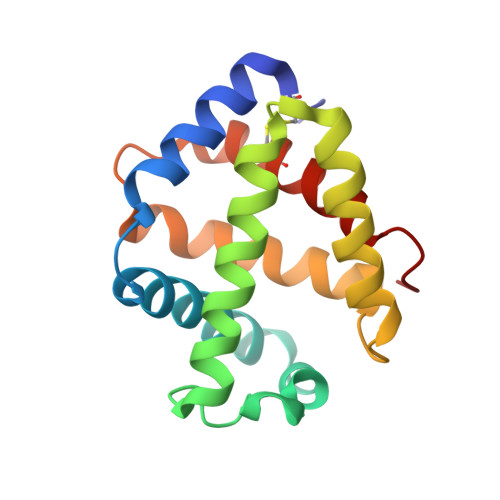
Sulfide binding is mediated by zinc ions discovered in the crystal structure of a hydrothermal vent tubeworm hemoglobin.
Flores, J.F., Fisher, C.R., Carney, S.L., Green, B.N., Freytag, J.K., Schaeffer, S.W., Royer Jr, W.E.(2005) Proc Natl Acad Sci U S A 102: 2713-2718
- PubMed: 15710902
- DOI: https://doi.org/10.1073/pnas.0407455102
- Primary Citation of Related Structures:
1YHU - PubMed Abstract:
Key to the remarkable ability of vestimentiferan tubeworms to thrive in the harsh conditions of hydrothermal vents are hemoglobins that permit the sequestration and delivery of hydrogen sulfide and oxygen to chemoautotrophic bacteria. Here, we demonstrate that zinc ions, not free cysteine residues, bind sulfide in vestimentiferan hemoglobins. The crystal structure of the C1 hemoglobin from the hydrothermal vent tubeworm Riftia pachyptila has been determined to 3.15 A and revealed the unexpected presence of 12 tightly bound Zn(2+) ions near the threefold axes of this D(3) symmetric hollow sphere. Chelation experiments on R. pachyptila whole-coelomic fluid and purified hemoglobins reveal a role for Zn(2+) ions in sulfide binding. Free cysteine residues, previously proposed as sulfide-binding sites in vestimentiferan hemoglobins, are found buried in surprisingly hydrophobic pockets below the surface of the R. pachyptila C1 molecule, suggesting that access of these residues to environmental sulfide is restricted. Attempts to reduce the sulfide-binding capacities of R. pachyptila hemoglobins by addition of a thiol inhibitor were also unsuccessful. These findings challenge the currently accepted paradigm of annelid hemoglobin evolution and adaptation to reducing environments.
Organizational Affiliation:
Department of Biology, Pennsylvania State University, University Park, PA 16802, USA. jff133@psu.edu