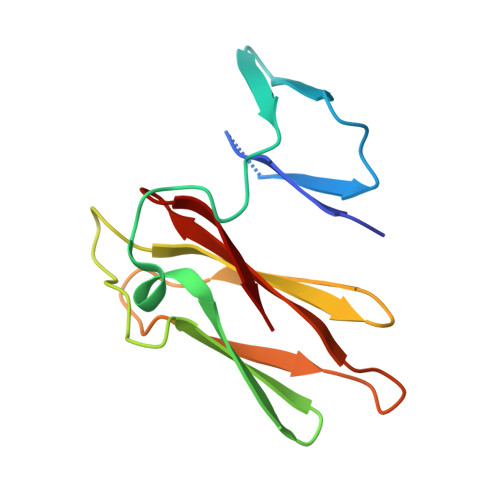
The first crystal structure of class III superoxide reductase from Treponema pallidum.
Santos-Silva, T., Trincao, J., Carvalho, A.L., Bonifacio, C., Auchere, F., Raleiras, P., Moura, I., Moura, J., Romao, M.J.(2006) J Biol Inorg Chem 11: 548-558
- PubMed: 16791639
- DOI: https://doi.org/10.1007/s00775-006-0104-y
- Primary Citation of Related Structures:
1Y07 - PubMed Abstract:
Superoxide reductase (SOR) is a metalloprotein containing a non-heme iron centre, responsible for the scavenging of superoxide radicals in the cell. The crystal structure of Treponema pallidum (Tp) SOR was determined using soft X-rays and synchrotron radiation. Crystals of the oxidized form were obtained using poly(ethylene glycol) and MgCl2 and diffracted beyond 1.55 A resolution. The overall architecture is very similar to that of other known SORs but TpSOR contains an N-terminal domain in which the desulforedoxin-type Fe centre, found in other SORs, is absent. This domain conserves the beta-barrel topology with an overall arrangement very similar to that of other SOR proteins where the centre is present. The absence of the iron ion and its ligands, however, causes a decrease in the cohesion of the domain and some disorder is observed, particularly in the region where the metal would be harboured. The C-terminal domain exhibits the characteristic immunoglobulin-like fold and harbours the Fe(His)4(Cys) active site. The five ligands of the iron centre are well conserved despite some disorder observed for one of the four molecules in the asymmetric unit. The participation of a glutamate as the sixth ligand of some of the iron centres in Pyrococcus furiosus SOR was not observed in TpSOR. A possible explanation is that either X-ray photoreduction occurred or there was a mixture of redox states at the start of data collection. In agreement with earlier proposals, details in the TpSOR structure also suggest that Lys49 might be involved in attraction of superoxide to the active site.
Organizational Affiliation:
REQUIMTE/CQFB, Departamento de Química, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa, 2829-516, Caparica, Portugal.