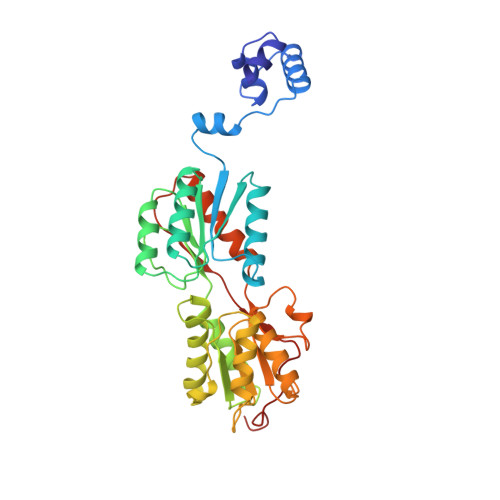
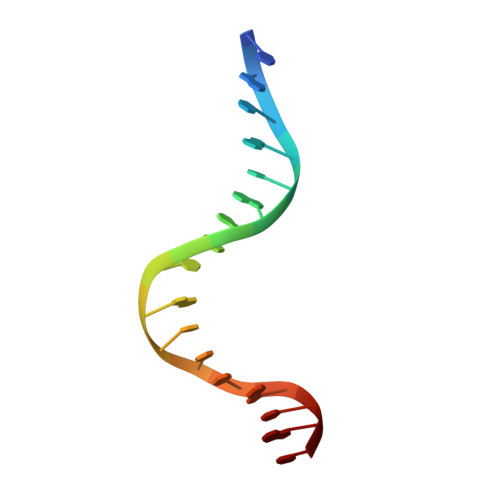
The structure of PurR mutant L54M shows an alternative route to DNA kinking.
Arvidson, D.N., Lu, F., Faber, C., Zalkin, H., Brennan, R.G.(1998) Nat Struct Biol 5: 436-441
- PubMed: 9628480
- DOI: https://doi.org/10.1038/nsb0698-436
- Primary Citation of Related Structures:
1VPW - PubMed Abstract:
The crystal structure of the purine repressor mutant L54M bound to hypoxanthine and to the purF operator provides a stereochemical understanding of the high DNA affinity of this hinge helix mutant. Comparison of the PurR L54M-DNA complex to that of the wild type PurR-DNA complex reveals that these purine repressors bind and kink DNA similarly despite significant differences in their minor groove contacts and routes to interdigitation of the central C.G:G.C base pair step. Modeling studies, supported by genetic and biochemical data, show that the stereochemistry of the backbone atoms of the abutting hinge helices combined with the rigidity of the kinked base pair step constrain the interdigitating residue to leucine or methionine for the LacI/GalR family of transcription regulators.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Oregon Health Sciences University, Portland 97201-3098, USA.