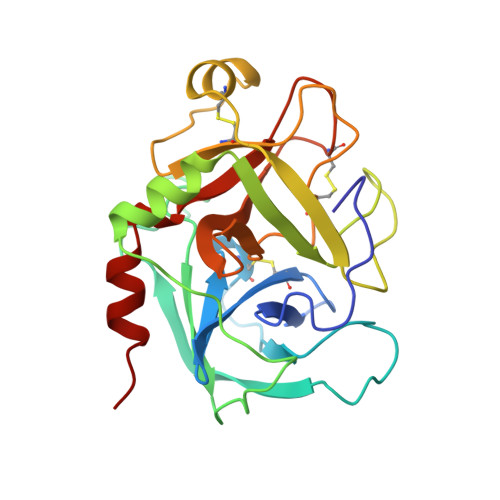
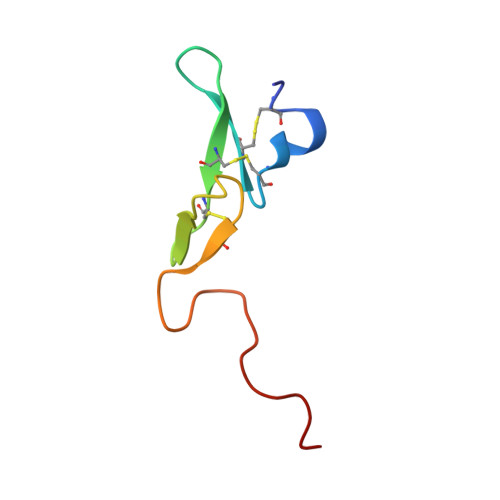
Coagulation factor IXa: the relaxed conformation of Tyr99 blocks substrate binding.
Hopfner, K.P., Lang, A., Karcher, A., Sichler, K., Kopetzki, E., Brandstetter, H., Huber, R., Bode, W., Engh, R.A.(1999) Structure 7: 989-996
- PubMed: 10467148
- DOI: https://doi.org/10.1016/s0969-2126(99)80125-7
- Primary Citation of Related Structures:
1RFN - PubMed Abstract:
Among the S1 family of serine proteinases, the blood coagulation factor IXa (fIXa) is uniquely inefficient against synthetic peptide substrates. Mutagenesis studies show that a loop of residues at the S2-S4 substrate-binding cleft (the 99-loop) contributes to the low efficiency. The crystal structure of porcine fIXa in complex with the inhibitor D-Phe-Pro-Arg-chloromethylketone (PPACK) was unable to directly clarify the role of the 99-loop, as the doubly covalent inhibitor induced an active conformation of fIXa.
Organizational Affiliation:
Abteilung Strukturforschung, Max-Planck-Institut für Biochemie, Martinsried, Germany.