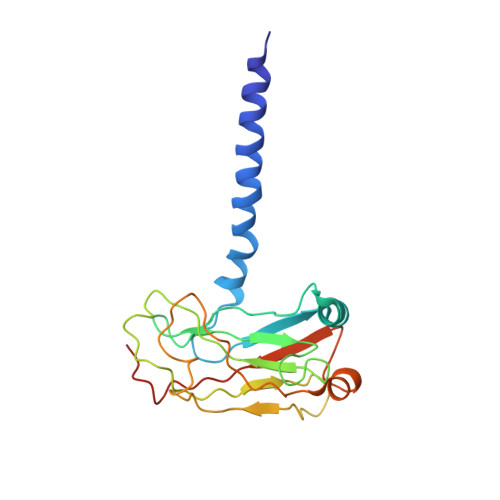
Structurally distinct recognition motifs in lymphotoxin-beta receptor and CD40 for tumor necrosis factor receptor-associated factor (TRAF)-mediated signaling.
Li, C., Norris, P.S., Ni, C.Z., Havert, M.L., Chiong, E.M., Tran, B.R., Cabezas, E., Reed, J.C., Satterthwait, A.C., Ware, C.F., Ely, K.R.(2003) J Biol Chem 278: 50523-50529
- PubMed: 14517219
- DOI: https://doi.org/10.1074/jbc.M309381200
- Primary Citation of Related Structures:
1RF3 - PubMed Abstract:
Lymphotoxin-beta receptor (LTbetaR) and CD40 are members of the tumor necrosis factor family of signaling receptors that regulate cell survival or death through activation of NF-kappaB. These receptors transmit signals through downstream adaptor proteins called tumor necrosis factor receptor-associated factors (TRAFs). In this study, the crystal structure of a region of the cytoplasmic domain of LTbetaR bound to TRAF3 has revealed an unexpected new recognition motif, 388IPEEGD393, for TRAF3 binding. Although this motif is distinct in sequence and structure from the PVQET motif in CD40 and PIQCT in the regulator TRAF-associated NF-kappaB activator (TANK), recognition is mediated in the same binding crevice on the surface of TRAF3. The results reveal structurally adaptive "hot spots" in the TRAF3-binding crevice that promote molecular interactions driving specific signaling after contact with LTbetaR, CD40, or the downstream regulator TANK.
Organizational Affiliation:
Cancer Research Center, The Burnham Institute, La Jolla, California 92037, USA.