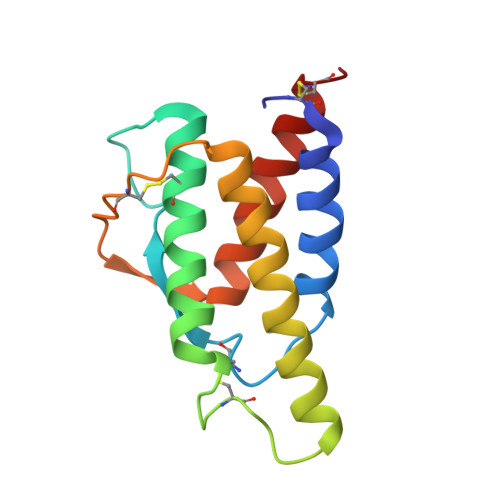
Crystal structure of human recombinant interleukin-4 at 2.25 A resolution.
Wlodawer, A., Pavlovsky, A., Gustchina, A.(1992) FEBS Lett 309: 59-64
- PubMed: 1511746
- DOI: https://doi.org/10.1016/0014-5793(92)80739-4
- Primary Citation of Related Structures:
1RCB - PubMed Abstract:
The crystal structure of human recombinant interleukin-4 (IL-4) has been solved by multiple isomorphous replacement, and refined to an R factor of 0.218 at 2.25 A resolution. The molecule is a left-handed four-helix bundle with a short stretch of beta sheet. The structure bears close resemblance to other cytokines such as granulocyte-macrophage colony stimulating factor (GM-CSF). Although no sequence similarity of IL-4 to GM-CSF and other related cytokines has been previously postulated, structure-based alignment of IL-4 and GM-CSF revealed that the core of the molecules, including large parts of all four helices and extending over half of the molecule, has 30% sequence identity. This may have identified regions which are not only important to maintain structure, but could also play a role in receptor binding.
Organizational Affiliation:
Macromolucar Structure Laboratory, NCI-Fredrick Cancer Research and Development Center, MD 21702.