Insights into Catalysis by a Knotted TrmD tRNA Methyltransferase.
Elkins, P.A., Watts, J.M., Zalacain, M., Van Thiel, A., Vitaszka, P.R., Redlak, M., Andraos-Selim, C., Rastinejad, F., Holmes, W.M.(2003) J Mol Biol 333: 931-949
- PubMed: 14583191
- DOI: https://doi.org/10.1016/j.jmb.2003.09.011
- Primary Citation of Related Structures:
1P9P - PubMed Abstract:
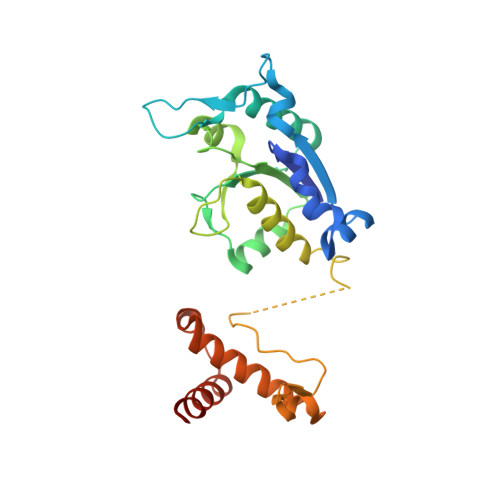
The crystal structure of Escherichia coli tRNA (guanosine-1) methyltransferase (TrmD) complexed with S-adenosyl homocysteine (AdoHcy) has been determined at 2.5A resolution. TrmD, which methylates G37 of tRNAs containing the sequence G36pG37, is a homo-dimer. Each monomer consists of a C-terminal domain connected by a flexible linker to an N-terminal AdoMet-binding domain. The two bound AdoHcy moieties are buried at the bottom of deep clefts. The dimer structure appears integral to the formation of the catalytic center of the enzyme and this arrangement strongly suggests that the anticodon loop of tRNA fits into one of these clefts for methyl transfer to occur. In addition, adjacent hydrophobic sites in the cleft delineate a defined pocket, which may accommodate the GpG sequence during catalysis. The dimer contains two deep trefoil peptide knots and a peptide loop extending from each knot embraces the AdoHcy adenine ring. Mutational analyses demonstrate that the knot is important for AdoMet binding and catalytic activity, and that the C-terminal domain is not only required for tRNA binding but plays a functional role in catalytic activity.
Organizational Affiliation:
GlaxoSmithKline, 709 Swedeland Road, UE0447, King of Prussia, PA 19406, USA.