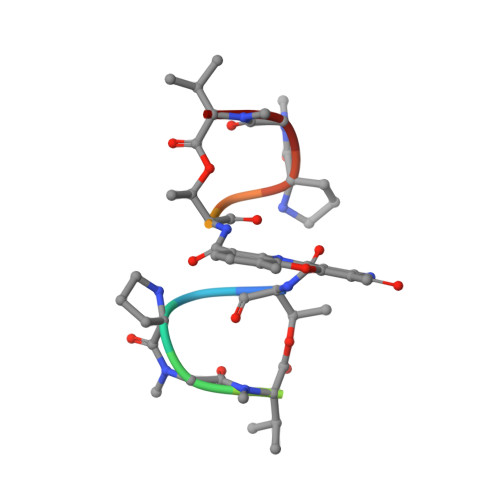
Solution Structure of the Actd-5'-Ccgtt(3)Gtgg-3' Complex: Drug Interaction with Tandem G.T Mismatches and Hairpin Loop Backbone.
Chin, K.-H., Chen, F.-M., Chou, S.-H.(2003) Nucleic Acids Res 31: 2622
- PubMed: 12736312
- DOI: https://doi.org/10.1093/nar/gkg353
- Primary Citation of Related Structures:
1OVF - PubMed Abstract:
Binding of actinomycin D (ActD) to the seemingly single-stranded DNA (ssDNA) oligomer 5'-CCGTT3 GTGG-3' has been studied in solution using high-resolution nuclear magnetic resonance (NMR) techniques. A strong binding constant (8 x 10(6) M(-1)) and high quality NMR spectra have allowed us to determine the initial DNA structure using distance geometry as well as the final ActD-5'-CCGTT3 GTGG-3' complex structure using constrained molecular dynamics calculations. The DNA oligomer 5'-CCGTT3GTGG-3' in the complex forms a hairpin structure with tandem G.T mismatches at the stem region next to a loop of three stacked thymine bases pointing toward the major groove. Bipartite T2O-GH1 and T2O-G2NH2 hydrogen bonds were detected for the G.T mismatches that further stabilize this unusual DNA hairpin. The phenoxazone chromophore of ActD intercalates nicely between the tandem G.T mismatches in essentially one major orientation. Additional hydrophobic interactions between the ActD quinoid amino acid residues with the loop T5-T6-T7 backbone protons were also observed. The hydrophobic G-phenoxazone-G interaction in the ActD-5'-CCGTT3GTGG-3' complex is more robust than that of the classical ActD- 5'-CCGCT3GCGG-3' complex, consistent with the roughly 2-fold stronger binding of ActD to the 5'-CCGTT3GTGG-3' sequence than to its 5'-CCG CT3GCGG-3' counterpart. Stabilization by ActD of a hairpin containing non-canonical stem base pairs further strengthens the notion that ActD or other related compounds may serve as a sequence- specific ssDNA-binding agent that inhibits human immunodeficiency virus (HIV) and other retroviruses replicating through ssDNA intermediates.
Organizational Affiliation:
Institute of Biochemistry, National Chung-Hsing University, Taichung, 40227, Taiwan.