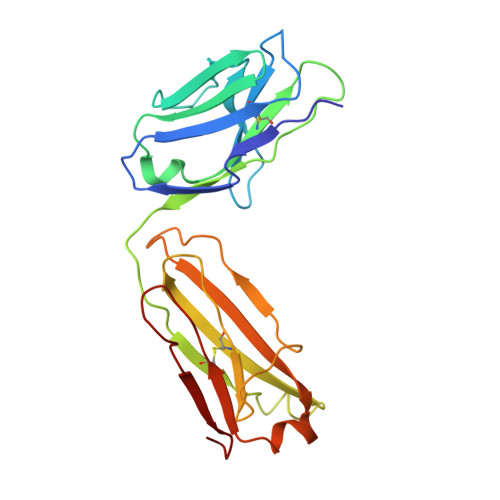
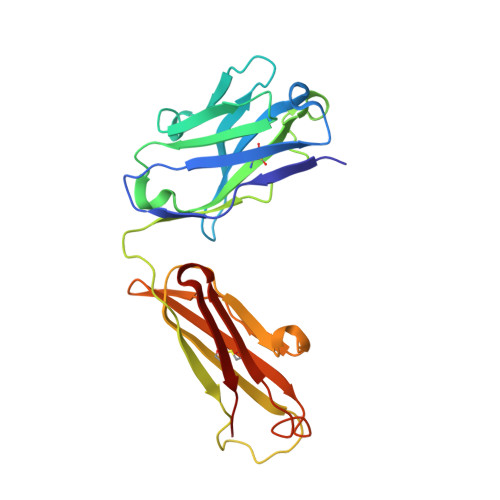
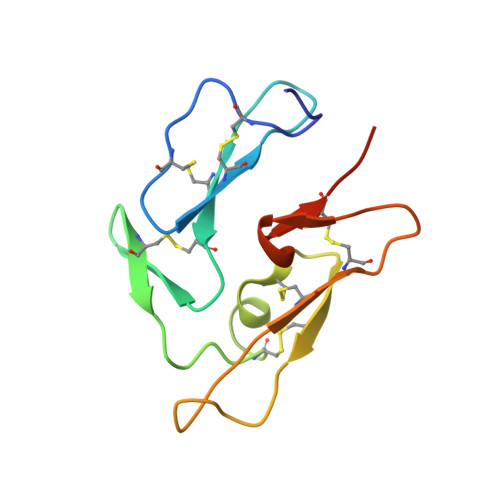
Crystal Structure of a Fab Complex Formed with Pfmsp1-19, the C-Terminal Fragment of Merozoite Surface Protein 1 from Plasmodium Falciparum: A Malaria Vaccine Candidate
Pizarro, J.C., Chitarra, V., Verger, D., Holm, I., Petres, S., Dartville, S., Nato, F., Longacre, S., Bentley, G.A.(2003) J Mol Biol 328: 1091
- PubMed: 12729744
- DOI: https://doi.org/10.1016/s0022-2836(03)00376-0
- Primary Citation of Related Structures:
1OB1 - PubMed Abstract:
Merozoite surface protein 1 (MSP1) is the major protein component on the surface of the merozoite, the erythrocyte-invasive form of the malaria parasite Plasmodium. Present in all species of Plasmodium, it undergoes two distinct proteolytic maturation steps during the course of merozoite development that are essential for invasion of the erythrocyte. Antibodies specific for the C-terminal maturation product, MSP1-19, can inhibit erythrocyte invasion and parasite growth. This polypeptide is therefore considered to be one of the more promising malaria vaccine candidates. We describe here the crystal structure of recombinant MSP1-19 from P.falciparum (PfMSP1-19), the most virulent species of the parasite in humans, as a complex with the Fab fragment of the monoclonal antibody G17.12. This antibody recognises a discontinuous epitope comprising 13 residues on the first epidermal growth factor (EGF)-like domain of PfMSP1-19. Although G17.12 was raised against the recombinant antigen expressed in an insect cell/baculovirus system, it binds uniformly to the surface of merozoites from the late schizont stage, showing that the cognate epitope is exposed on the naturally occurring MSP1 polypeptide complex. Although the epitope includes residues that have been mapped to regions recognised by invasion-inhibiting antibodies studied by other workers, G17.12 does not inhibit erythrocyte invasion or MSP1 processing.
Organizational Affiliation:
Unité d'Immunologie Structurale (CNRS URA 2185), Département de Biologie Structurale et Chimie, Institut Pasteur, 25 rue du Dr. Roux, 75724 Paris, cedex 15, France.