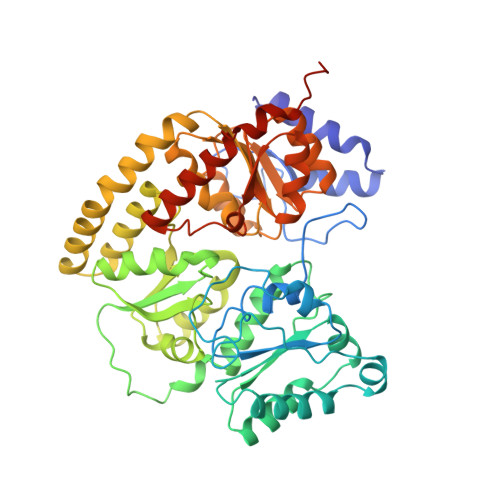
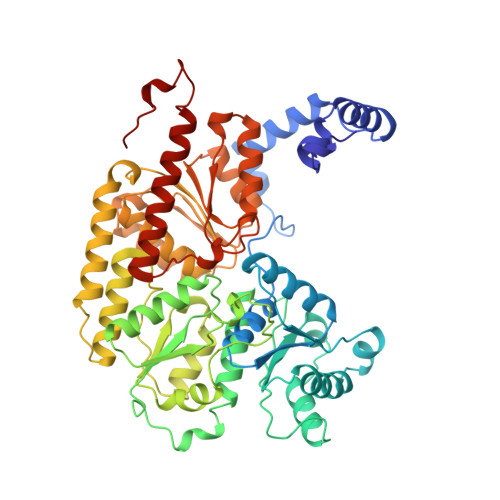
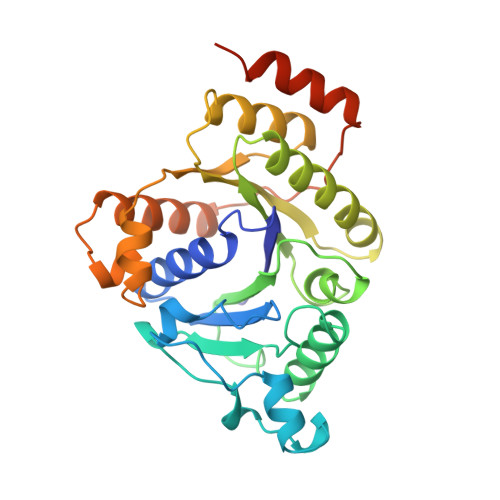
Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction.
Schindelin, H., Kisker, C., Schlessman, J.L., Howard, J.B., Rees, D.C.(1997) Nature 387: 370-376
- PubMed: 9163420
- DOI: https://doi.org/10.1038/387370a0
- Primary Citation of Related Structures:
1N2C - PubMed Abstract:
The coupling of ATP hydrolysis to electron transfer by the enzyme nitrogenase during biological nitrogen fixation is an important example of a nucleotide-dependent transduction mechanism. The crystal structure has been determined for the complex between the Fe-protein and MoFe-protein components of nitrogenase stabilized by ADP x AIF4-, previously used as a nucleoside triphosphate analogue in nucleotide-switch proteins. The structure reveals that the dimeric Fe-protein has undergone substantial conformational changes. The beta-phosphate and AIF4- groups are stabilized through intersubunit contacts that are critical for catalysis and the redox centre is repositioned to facilitate electron transfer. Interactions in the nitrogenase complex have broad implications for signal and energy transduction mechanisms in multiprotein complexes.
Organizational Affiliation:
Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena 91125, USA.