Structure/function insights into Tn5 transposition.
Steiniger-White, M., Rayment, I., Reznikoff, W.S.(2004) Curr Opin Struct Biol 14: 50-57
- PubMed: 15102449
- DOI: https://doi.org/10.1016/j.sbi.2004.01.008
- Primary Citation of Related Structures:
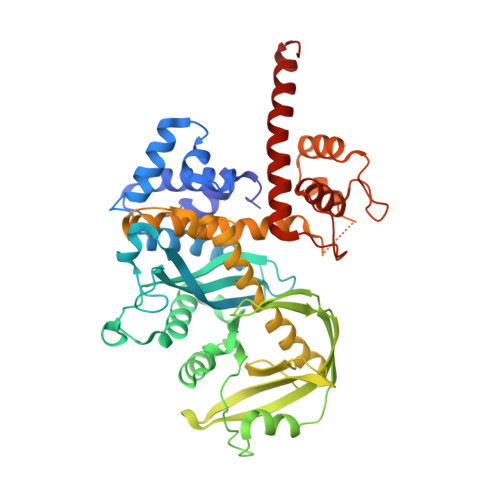
1MUS - PubMed Abstract:
Prokaryotic transposon 5 (Tn5) serves as a model system for studying the molecular mechanism of DNA transposition. Elucidation of the X-ray co-crystal structure of Tn5 transposase complexed with a DNA recognition end sequence provided the first three-dimensional picture of an intermediate in a transposition/retroviral integration pathway. The many Tn5 transposase-DNA co-crystal structures now available complement biochemical and genetic studies, allowing a comprehensive and detailed understanding of transposition mechanisms. Specifically, the structures reveal two different types of protein-DNA contacts: cis contacts, required for initial DNA recognition, and trans contacts, required for catalysis. Protein-protein contacts required for synapsis are also seen. Finally, the two divalent metals in the active site of the transposase support a 'two-metal-ion' mechanism for Tn5 transposition.
Organizational Affiliation:
University of Wisconsin-Madison, Department of Biochemistry, 433 Babcock Drive, Madison, WI 53706, USA. mmsteini@wisc.edu