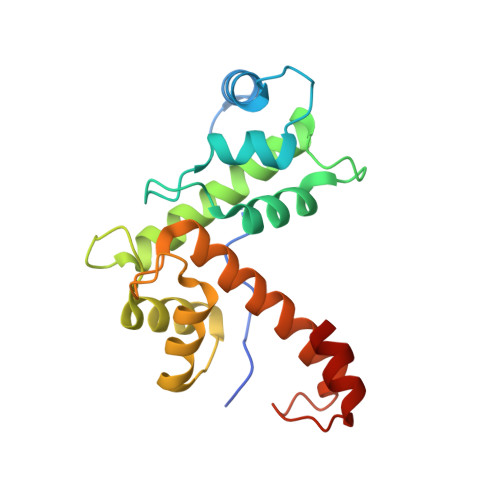
Structure of apoptosis-linked protein ALG-2: insights into Ca2+-induced changes in penta-EF-hand proteins.
Jia, J., Tarabykina, S., Hansen, C., Berchtold, M., Cygler, M.(2001) Structure 9: 267-275
- PubMed: 11525164
- DOI: https://doi.org/10.1016/s0969-2126(01)00585-8
- Primary Citation of Related Structures:
1HQV - PubMed Abstract:
The Ca2+ binding apoptosis-linked gene-2 (ALG-2) protein acts as a proapoptotic factor in a variety of cell lines and is required either downstream or independently of caspases for apoptosis to occur. ALG-2 belongs to the penta-EF-hand (PEF) protein family and has two high-affinity and one low-affinity Ca2+ binding sites. Like other PEF proteins, its N terminus contains a Gly/Pro-rich segment. Ca2+ binding is required for the interaction with the target protein, ALG-2 interacting protein 1 (AIP1).
Organizational Affiliation:
Biotechnology Research Institute, National Research Council of Canada, Montreal, Quebec.