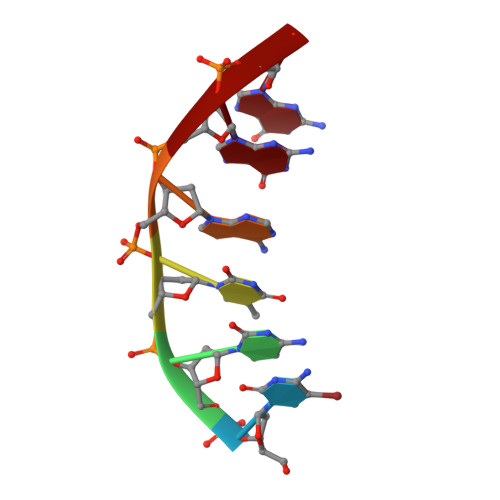
A DNA-porphyrin minor-groove complex at atomic resolution: the structural consequences of porphyrin ruffling.
Bennett, M., Krah, A., Wien, F., Garman, E., McKenna, R., Sanderson, M., Neidle, S.(2000) Proc Natl Acad Sci U S A 97: 9476-9481
- PubMed: 10920199
- DOI: https://doi.org/10.1073/pnas.160271897
- Primary Citation of Related Structures:
1EM0 - PubMed Abstract:
The crystal structure of a B-type DNA hexanucleotide duplex complexed with the porphyrin molecule nickel-[tetra-N-methyl-pyridyl] porphyrin has been solved by multiwavelength anomalous diffraction phasing and refined to an R factor of 11.5% at a resolution of 0.9 A. The structure has been solved and refined as two crystallographically independent duplexes, stacked end to end. Contrary to expectation, the porphyrin molecule is not intercalated into the duplex but is stacked onto the ends of the two-duplex stack. The porphyrin molecule is highly buckled as a consequence of the nickel coordination, which produces large changes in local DNA structure. A second mode of porphyrin binding is apparent as a consequence of crystal packing, which places the ligand in the minor groove of an adjacent duplex. This structure thus provides, to our knowledge, the first atomic visualization of minor-groove binding for a porphyrin molecule. The geometry of groove binding provides a ready explanation for porphyrin-induced DNA strand cleavage at deoxyribose residues.
Organizational Affiliation:
The Randall Institute, Department of Biomedical Sciences, King's College London, United Kingdom.